A brief introduction to PREDATOR
A brief introduction to PREDATORPREDATOR is a secondary structure prediction method based on recognition of potentially hydrogen-bonded residues in a single amino acid sequence (original server).
 A brief introduction to PREDATOR
A brief introduction to PREDATOR
PREDATOR is a secondary structure prediction method based on recognition of potentially hydrogen-bonded residues in a single
amino acid sequence (original server).
 Availability in NPS@
Availability in NPS@
This method is available :
 Parameters
Parameters
The user can choose between two database files stride.dat and dssp.dat. These files contain propensity tables, secondary structural
assignments and thresholds for two secondary structure assignment methods from tertiary structure : STRIDE and DSSP.
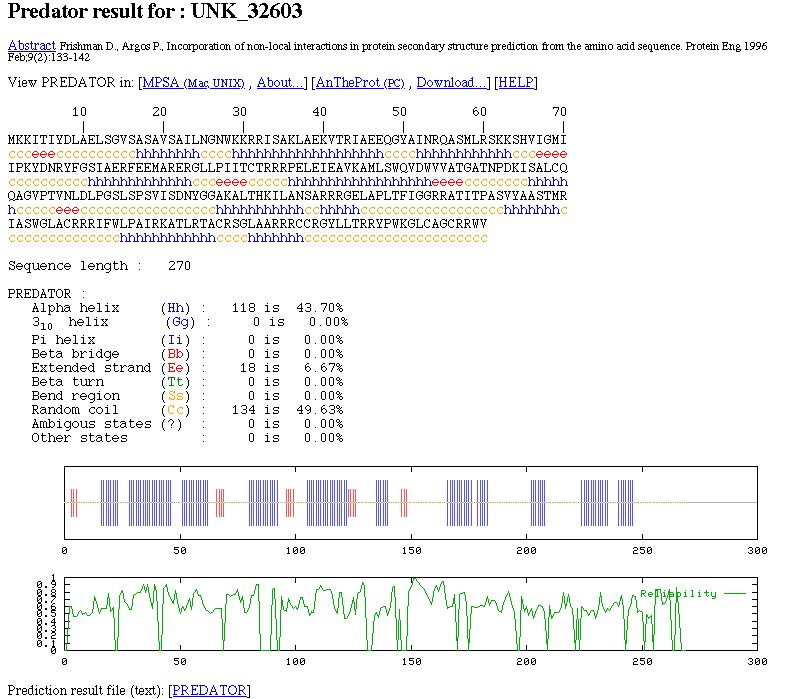
 NPS@ PREDATOR output example
NPS@ PREDATOR output example
You can see: