HNN help
 A brief introduction to HNN
A brief introduction to HNN
The HNN (Hierarchical Neural Network ) prediction method can be seen as an improvement on the famous classifier developed by Qian and
Sejnowski, and derived from the system NETtalk. As its predecessor, it is made up of two networks: a sequence-to-structure network and
a structure-to-structure network. The prediction is thus only based on local information. The improvements mainly deal with two points:
- technical tricks (recurrent connections, shared weights...) have been used to increase the context on which the prediction is made
and concomitantly decrease by two orders of magnitude the number of parameters (weights),
- physico-chemical data have been explicitly incorporated in the predictors used by the structure-to-structure network.
These modifications have significantly improved the error in generalization. This error could still be greatly lowered, would the
profile of multiple-alignment be used as input of the whole system, instead of the single primary structure of the sequence of
interest.
NPS@ is the orginal server for this method.
 Availability in NPS@
Availability in NPS@
This method is available :
 Parameters
Parameters
No parameter required.
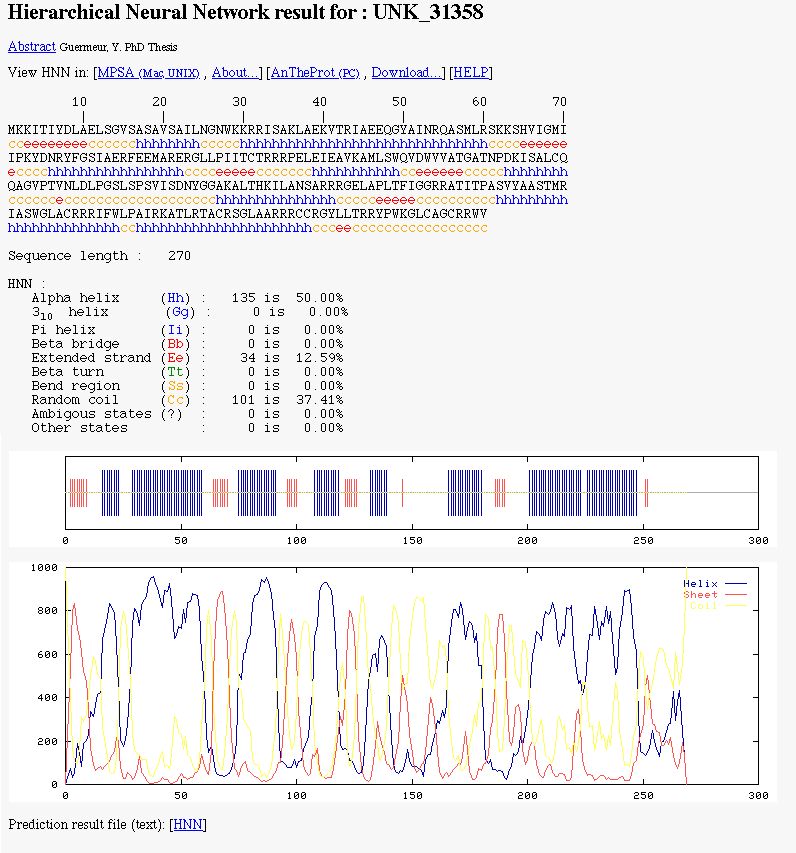
 NPS@ HNN output example
NPS@ HNN output example
You can see:
- MPSA/ANTHEPROT link to
view the prediction in these local protein sequence analysis
softwares.
- The color coded prediction (a sequence line and below the corresponding predicted states).
- The sequence length.
- The percentage of each secondary element.
- Two graphics. The first to better visualize the prediction. In the second, there are the score curves for each predicted
state.
- Links on the prediction result text file.
 References
References
 A brief introduction to HNN
A brief introduction to HNN