DSC help
 A brief introduction to DSC
A brief introduction to DSC
DSC (Discrimination of protein Secondary structure Class) is based on decomposing secondary structure prediction into the basic concepts
and then use of simple and linear statistical methods to combine the concepts for prediction. This makes the prediction method
comprehensible and allows the relative importance of the different sources of information used to be measured.
In NPS@, we run BLASTP on SWISS-PROT, filter it and then run CLUSTALW. The resulting alignment is the input for
DSC.
 Availability in NPS@
Availability in NPS@
This method is available :
 Parameters
Parameters
No parameter required.
 NPS@ DSC output example
NPS@ DSC output example
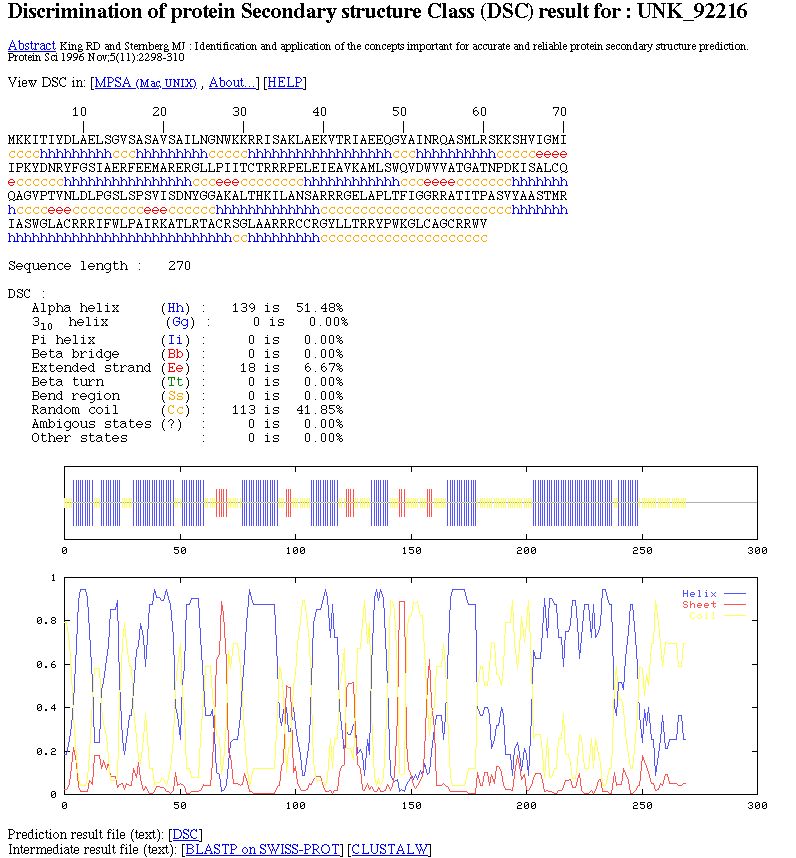
You can see:
- MPSA link to
view the prediction in this local protein sequence analysis
software.
- The color coded prediction (a sequence line and below the corresponding predicted states).
- The sequence length.
- The percentage of each secondary element.
- Two graphics. The first to better visualize the prediction. In the second, there are the score curves for each predicted
state.
- Links on the prediction result text file and on intermediate result text files.
 References
References
 A brief introduction to DSC
A brief introduction to DSC