DPM help
 A brief introduction to DPM
A brief introduction to DPM
DPM (Double Prediction Method) is a secondary structure prediction which predicts the protein structural class and then the
secondary structure for the sequence (this is why Double). The DPM method consists in 4 successive steps:
- Prediction of the structural class of a protein from AA composition (Nakashima et al. 1986 J. Biochem. Tokyo, 99, 153-162).
- Preliminary secondary structure estimation from a simple algorithm.
- Comparison between the 2 independent predictions.
- Optimization of parameters and reprediction of secondary structure.
NPS@ is the orginal server for this method.
 Availability in NPS@
Availability in NPS@
This method is available :
 Parameters
Parameters
No parameter required.
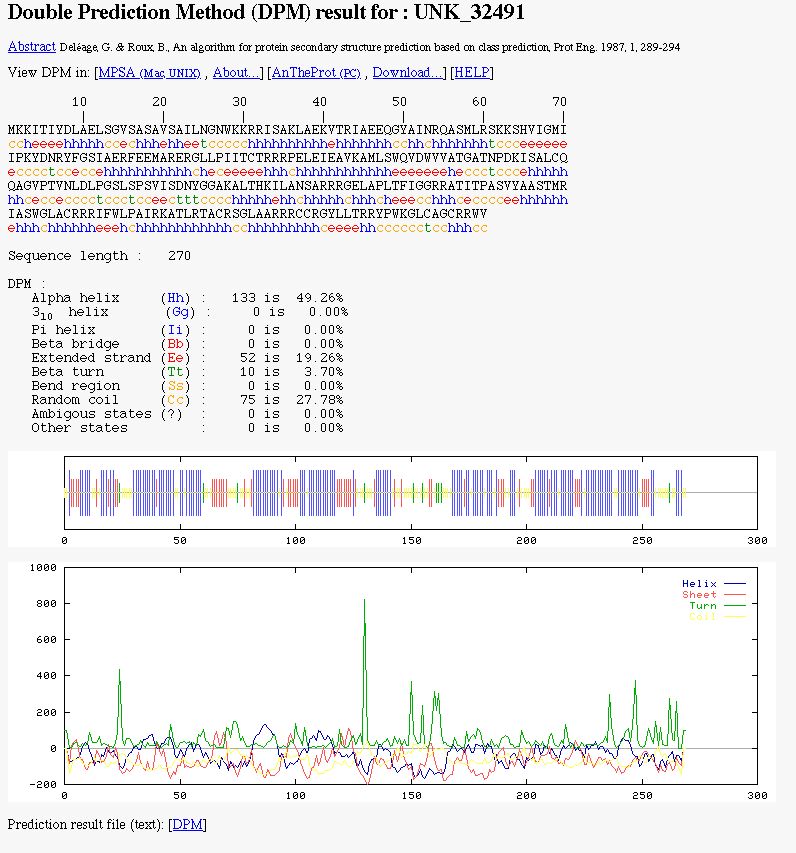
 NPS@ DPM output example
NPS@ DPM output example
You can see:
- MPSA/ANTHEPROT link to
view the prediction in these local protein sequence analysis
softwares.
- The color coded prediction (a sequence line and below the corresponding predicted states).
- The sequence length.
- The percentage of each secondary element.
- Two graphics. The first to better visualize the prediction. In the second, there are the score curves for each predicted
state.
- Links on the prediction result text file.
 References
References
 A brief introduction to DPM
A brief introduction to DPM