PFSEARCH help
 A brief introduction to PFSEARCH
A brief introduction to PFSEARCH
The program PFSEARCH, belonging to PFTOOLS package, compares a query PROSITE profile (generalized profile) against a sequence library. This is
a useful tool for homology detection, for the detection of motifs in protein sequences...
For further details about generalized profiles and PFTOOLS package,
see the ISREC Profile Homepage.
 Availability in NPS@
Availability in NPS@
PFSEARCH is available :
 Parameters
Parameters
The searching options correspond to the choice of the protein library and to the choice of the disjointness mode (with unique
mode, multiple profile-sequence alignment between the same profile and the same sequence are not permitted).
In NPS@, pfsearch is used with normalized scores. Consequently, you have to choose a cut-off value for normalized scores.
 NPS@ PFSEARCH output example
NPS@ PFSEARCH output example
The NPS@ PFSEARCH output is divided into four parts.
-
PART 1:
- First, you have a form to build an alignment from matches found by PFSEARCH. The alignment you will generate, corresponds to a
sub-alignment of the master alignment which has been obtained by aligning all matches found to the profil. To build your alignment, you can :
- select sequences with a normalized score upper or equal to a value of your choice.
By default, only sequences with a normalized score upper or equal to 8.5 are selected.
- choose an alignment region relatively to the master alignment.
- Your choices are validated when you click on the REDISPLAY button.
- If you want to come back to the original selections, then click on the RESET button.

-
PART 2:
This part corresponds to the list of matches found by PFSEARCH. Theses matches are sorted by order of score.
For each match, you can see :
- A checkbox to select/unselect sequence.
- The NPSA link allows you to apply
NPS@ methods on the corresponding sequence after it is extracted from the query database.
- The database link to retrieve the database entry.
- The length match which corresponds to the length of sequence fragment matching the profile.
- The normalized score.
- The number of displayed sequences.
- The number of selected sequences.

-
PART 3:
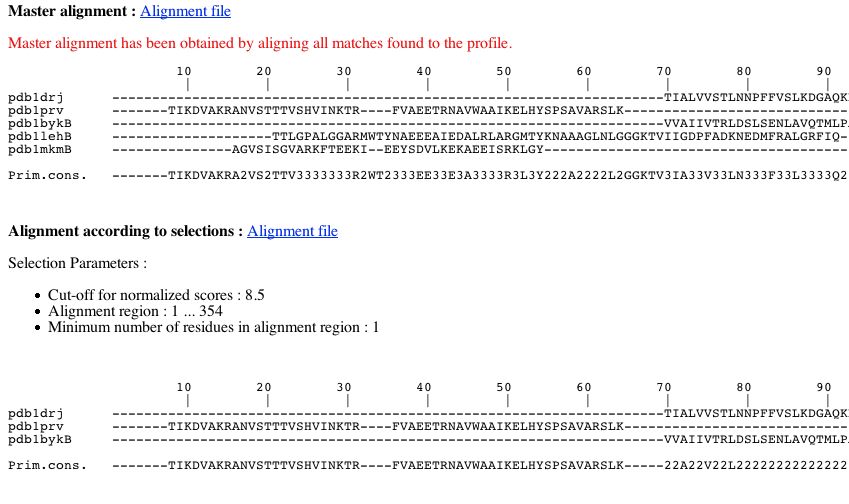
You can see two alignments:
- The master alignment obtained by aligning all matches found to the profile. Consequently this alignment can present columns with only gaps.
- The alignment according to your selections. As said previously, it corresponds to a sub-alignment of the master alignment :
- this alignment is composed from sequences whose normalised score is upper or equal to threshold chosen previously.
- this alignment corresponds to a particular region of the master alignment according to boundaries chosen in PART 1.
-
PART 4:
You have :
- A PFMAKE form to follow your analysis by using PFMAKE.
The alignment which will be used by PFMAKE corresponds to your last selections. Columns containing only gaps
will be removed. So be careful about the consistency of the generated alignment.
A checkbox allows you to remove identical sequences in the created alignment.
- An EXTRACT form to extract selected sequences and make a database. You can extract full or partial sequences.
The extraction is done on your current selection.
The full extract is made from the database.
The partial extract is made from selected sequences which contain a given number of residues in the indicated alignment region.
The extracted sequences are retrieved from the PFSEARCH alignment.
A checkbox allows you to remove identical sequences in the created database.
- A link on the original PFSEARCH result text file.

 References
References
 A brief introduction to PFSEARCH
A brief introduction to PFSEARCH


