PATTINPROT help
 A brief introduction to PATTINPROT
A brief introduction to PATTINPROT
PATTINPROT is a tool to scan a protein database of one or several sequences for one or several patterns. The pattern must
be written with the PROSITE syntax. PATTINPROT allows errors
towards pattern to find. Errors are set by number of mistmatch allowed or by a similarity threshold towards the pattern.
Algorithm :
In the the algorithm used by PATTINPROT and PROSCAN, the biological information is variably distributed between the pattern elements.
To do that, when an element is not respected, it's penalized by its frequency. When a strict element (e.g. -F- ) is not respected,
it's a heavy penalty cost for the search. So, the signal-noise ratio is improved.
This algorithm was developped in our laboratory and is included (NPS@ is the original server for it).
 Availability in NPS@
Availability in NPS@
PATTINPROT is available :
So, for example, you can use PATTINPROT to filter a database built
from an ACNUC query or
after a BLAST search.
 Parameters
Parameters
By default the scan is made with no error.
You can set the number of residues to show on both sides of the hit.
 NPS@ PATTINPROT output example
NPS@ PATTINPROT output example
The NPS@ PATTINPROT output is divided into three parts.
-
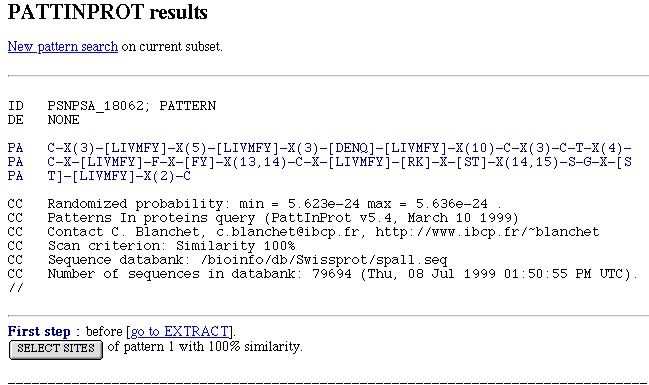
PART 1:
In this part, you have :
- A link to make a new pattern search on the current subset. The current subset include all sequences containing all
patterns searched since the start of the PATTINPROT search.
- The pattern definition.
- A form to select pattern. You have one button per pattern search. When you click on a button corresponding to a pattern,
all other patterns are unselected and you can only work with the sites corresponding to the button that you click on.
-
PART 2:
In this part you can found the sites found by PATTINPROT for each sequence of the database.
You can see :
- On the first line :
- The NPSA link allowing you to work with the sequence.
- A link to retrieve the database entry.
- A comment on the sequence (if any).
The links are active if the databank identifier (before the first |) is recognized by NPS@.
- For each site found :
- A checkbox to select/unselect the site.
- The site position within the sequence.
- The similarity level between the matching site and the query pattern.
- The site sequence in upper-case letters surrounded by '_' and the extra-residues (in lower-case
letters) if any and as set in parameters.

-
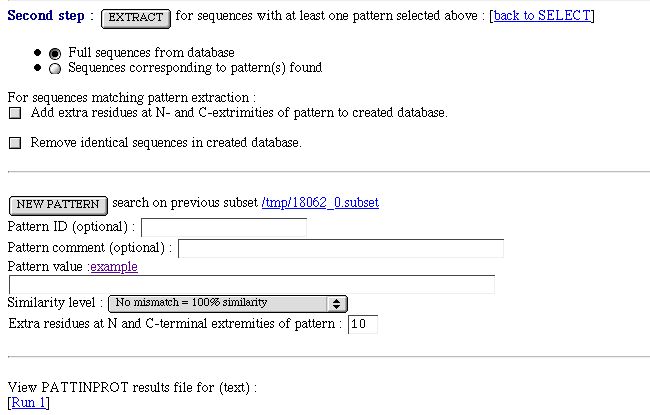
PART 3:
You have :
- An extract form to make a sub-database and work with
it.
You can then extract:
- full sequence from the database. This, for sequence with at least one site selected.
- the sequence of the matching site.
A checkbox to add residues at N- and C- terminal extrimities (for the sequence matching site extraction).
A checkbox to remove identical sequences in the created
database.
- The new pattern search form. The search is done on the subset database created after the previous PATTINPROT runs. All
sequences in the subset have all the previous search pattern.
 References
References
 A brief introduction to PATTINPROT
A brief introduction to PATTINPROT

