FIT help
 A brief introduction to FIT
A brief introduction to FIT
FIT is a program devoted to the 3D structure superimposition of molecules after sequence alignment. The basic algorithm for fitting molecules was kindly provided by P. Koehl in Strasbourg. The program has been extented by C. Geourjon in order to allow amino acid selection. The perl interface was written by F. David.
The FIT program is currently limited to the superimposition of up to 25 chains.
The logical flow chart is as follows :
- Upload PDB file names (i.e.1cdo:3bto) : Only PDB files can be used as input files or upload your personal PDB files
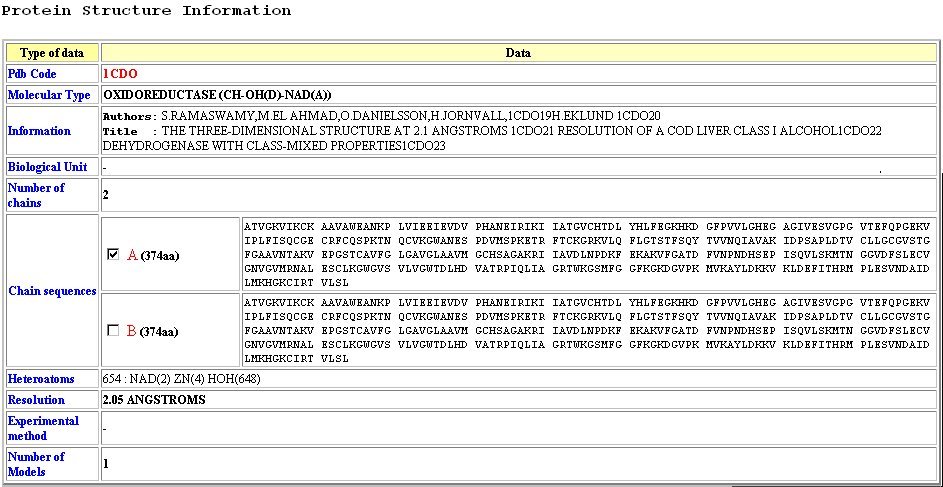
- Display PDB files information:
- Pdb Code Standard pdb entry code 1cdo:3bto
- Molecular Type
- Information Structure authors and title
- Biological Unit Biological information for oligomerisation state
- Number of chains Number of chain
- Chain sequences Sequence of each detected chain in the PDB file. Only X at the beginning or at the end of the sequence are removed
- Heteroatoms Provide information about number and nature of heteroatoms (number : heretoatom type)
- Resolution Only filled for X-ray structure
- Experimental method X-ray, NMR or -
- Number of Models Indicates the number of models if method used is NMR
- Chain selection : Obtained by selection buttons (Select all) or (Unselect all) or by checkboxes (one per sequence)
- Alignment : Align button CLUSTALW is used to perform the alignment.
- Amino acid selection : A list of amino acid with counterpart is available in a list allowing multiple selections.(amino acid facing gaps are not selectable)
- Superimposition of 3D structures using the chosen amino acid selection.
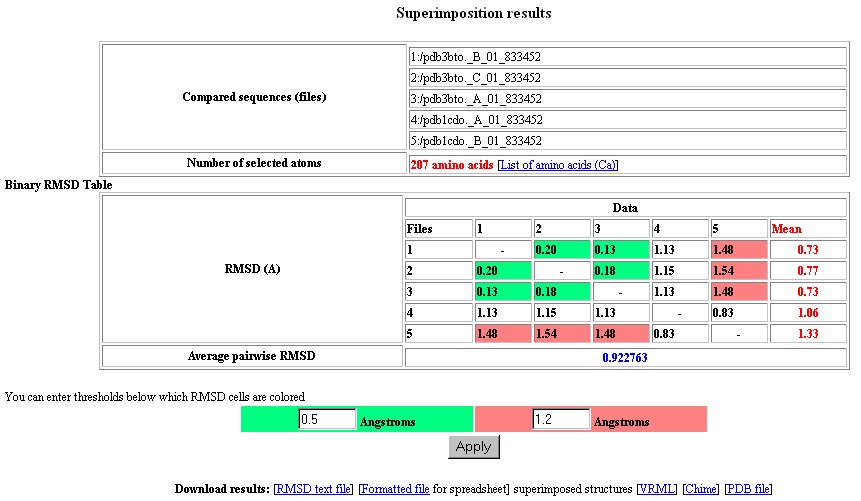
- Results display Table of binary fit with 2 coloring thresholds, and various links to files [VRML]
 Availability in NPS@
Availability in NPS@
This program is available :
 Parameters
Parameters
CLUSTALW parameters are not currently available for the user.
 NPS@ FIT output example 1cdo:3bto
NPS@ FIT output example 1cdo:3bto
The NPS@ FIT output is divided into 4 successive pages
-
Page 1:PDB files information
-
Page 2:Multiple alignment colored by using the CLUSTALW amino acid groups

-
Page 3:Display of binary fit values
-
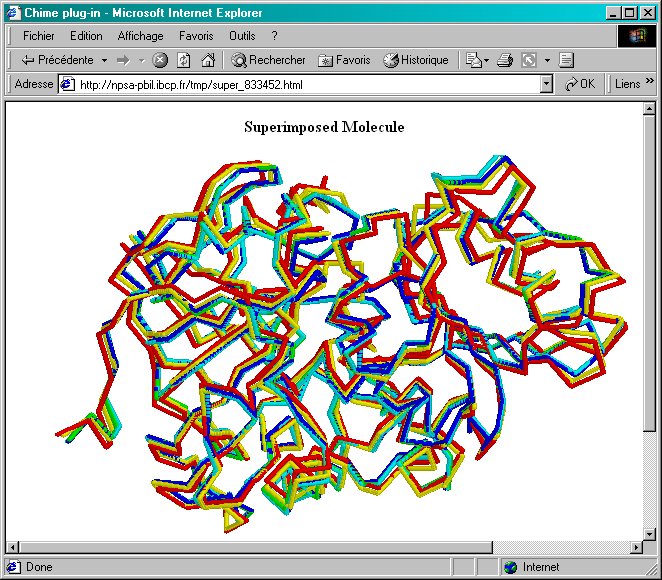
Page 4:Display of 3D superimposed structures. This display is available either with a VRML or a Chime plug-in.
 A brief introduction to FIT
A brief introduction to FIT


